RT-PCR Tests for COVID-19 Have Quantitative Power. Let’s Start Using It

Mara Aspinall, managing director, BlueStone Venture Partners; professor of the practice, biomedical diagnostics, Arizona State University
Over the past 16 months, 192 billion COVID tests have been carried out globally. A majority are real-time polymerase chain reaction tests (RTqPCR) tests.
Every one of those has been reported to patients as an either/or result: positive or, more usually, negative.
These tests are capable of doing much, much more than just giving a simple yes/no answer.
The “q” in RTqPCR measures “quantitative” viral load. We know this matters a lot — and every RTqPCR provides the “q” in terms of Ct value — how many cycles it took to detect a real signal from your samples.
Why do we systematically discard this data which is absolutely critical to both public health and clinical decisions?
Without an apples-to-apples standard to compare RTqPCR results, an individual with a high 3,000,000 copies/milliliter viral load will get the same positive lab result as another with a low 50 copies/ml. The clinical and public health consequences of these two results are definitively very different.
The value of quantitative readouts has become more clear in recent months. The higher the viral load, the higher the chance of serious disease, hospital admission, and transmission to others, non-pharmaceutical interventions (NPI) notwithstanding: physical distancing; mask wearing; well-ventilated spaces; quarantine; etc.
Simply put, the higher the viral load, the higher the chance of a patient ending up in the ICU.
The lower the viral load, the more likely the patient is not capable of viral transmission. Asymptomatic patients and those with only mild symptoms are less likely to transmit the virus to others because there is less virus to be broadcast in aerosolized respiratory droplets. All patients will be recorded as positive long after any likelihood of transmissible infection. Patients recovering from COVID-19 often remain PCR-positive for days or weeks, even up to six months, when the virus being detected may be only residual virus fragments.
The first evidence these patients may get is when they take a required PCR test two days before a flight — then they are surprised to find out they are “positive.”
There is consensus that frequent, while-you-wait community testing is best to inform both individual and public health actions. Traditional laboratory high-volume RTqPCR testing is automatically disqualified – it is too expensive to be used “frequently” and too slow to be “while-you-wait” (fastest results take 12-24 hours, and during the peak of the epidemic it stretched to 7-14 days – effectively useless except to inform an historical perspective).
Rapid antigen tests are cheap enough to be used frequently; and sensitive enough (~95%) to result positive in the period of highest viral load when individuals are infectious.
How do we judge what level of viral load is infectious?
Every infectious disease has a minimum quantity of virus necessary for an infection to take hold. This varies by virus and by an individual’s immune competency. The Lab gold standard to determine this is to infect a cell culture with SARS-CoV2 virus — these experiments indicate that anywhere from 1,000 to 100,000 viral copies per milliliter are required to create a viable COVID-19 infection.
There are caveats to these in-vitro lab tests – lab grown cell targets may not be representative of cells in patients; lab growth conditions are artificially optimized; there is no innate or adaptive immune reaction in a petri dish; etc.
However, this threshold is consistent with clinical experience — very few patients with moderate to severe disease sample below this level. (See this ASU T3 Blog from October 2020 “COVID-19 Test Accuracy – when is too much of a good thing bad?” for a fuller discussion of these issues and a bibliography.)
Why even perform RTqPCR tests at all?
Because the “q” is important in two ways: informing a patient’s clinical care; and if it can help identify individuals destined to become infectious early enough to pre-empt them from transmitting to others.
The key question is just how long is the pre-infectious stage?
Is it very small as shown below (green segment) (NEJM September 2020) or is it longer?
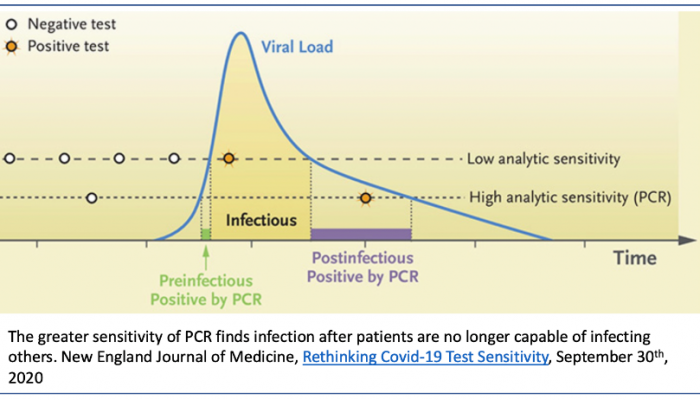
If the ramp up is fast and steep, the chance of any pre-infectious person receiving a traditional high sensitivity test quick enough is diminishingly low. Reliable data is hard to come by, since very few people are identified early enough to initiate the frequent repeat testing required to provide it.
Recent case reports from Caltech and the Pasadena Department of Public Health suggest that this detectable pre-infectious period can be as long as four days, especially among younger individuals. This implies there’s higher value to detecting individuals before they can infect others.
This finding reinforces the need for either: rapid antigen series testing (FDA announcement) or higher sensitivity (103 viral copies/ml or better) emerging point-of-care molecular tests (i.e. true PCR systems, more sensitive than most current POC LAMP systems).
PCR tests are exquisitely sensitive — the median test can detect 1 genome in a microliter of sample: the best are 100 times more sensitive; the worst 100 times less so, but even these are still able to detect the vast majority of infected individuals.
Every test reports a “q” — the cycle threshold (Ct, aka Cq, or Cp). However, the specifics of the test protocol affect what this Ct is — the same sample tested with different protocols will likely not generate comparable Ct numbers: it will vary protocol to protocol based on differences in pre-PCR processes (sample collection; use of transport medium; cDNA generation; reagent selection and purity); locations and base content of genome regions selected; primer design to bridge those regions (e.g. off-target binding or primer-dimer formation); probe design to detect amplified product; efficiency of the PCR cycler instrument used; etc.
Across all samples run on the same protocol, Ct will accurately reflect relative viral load differences sample to sample.
Generating comparability beyond that requires each lab to publish what is called a “Standard Curve” for each test protocol it performs. This translates “apples and oranges” Ct counts to more comparable viral loads expressed in terms of number of viral copies per milliliter.
This is done by taking a sample of known viral concentration (available commercially) and running a series of 10x dilutions on the same protocol and recording the resulted Ct with each known level of viral load — for that specific test, run in that particular way, by that particular lab.
As a demonstration we plotted Ct versus viral load for a more-or-less random group of 8 assays reported in the academic literature.
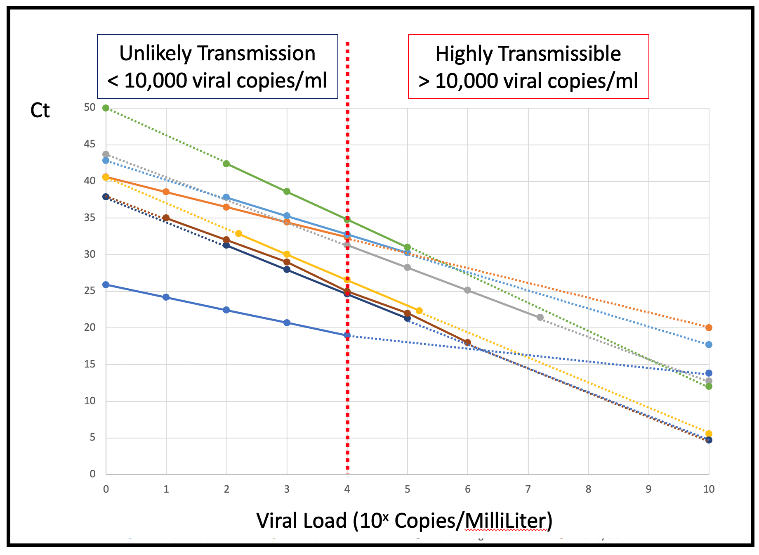
The vast majority of these have similar slopes – because they all use PCR which at 100% efficiency doubles the amplified product with each cycle. However, they have very different intercept Ct counts. A reported 20Ct for the most sensitive protocol implies a relatively less transmissible 1,000 to 10,000 (103-104) copies/ml in the sample.
While an apparently identical 20Ct for the least sensitive protocol means a highly infectious 10,000,000,000 (1010) copies/ml in the sample. At 100,000 (105) viral copies/ml Ct counts vary from 18Ct to 32Ct.
To talk of viral load only in Ct terms is both misleading and effectively meaningless beyond the bounds of any one individual assay protocol, unless the standard curve is created and Ct translated to viral load. (There still remain non-analytic issues that can erode comparability, for example: SARS-CoV-2 is tissue resident so the load available to sample from the respiratory tract may not directly reflect active virus driving clinical outcomes for the patient and their risk of transmissibility.)
All labs do calculate a standard curve as part of assay/instrument calibration for FDA-cleared, quantitative assays. However, this type of calibration is not required and rarely performed for qualitative assays, even if — as with RTqPCR — assays are inherently and robustly quantitative.
Current SARS-CoV-2 assays are approved by the FDA only for yes or no answers. The FDA has never before allowed reporting of a quantitative result from a qualitative viral test without requiring the calibrated standard curve in viral copies/ml on which the yes/no answer is based, and therefore allowing cross-assay comparison.
Even though the FDA hasn’t done this before, it’s easy to see how it could clear the way for this more quantitative view.
SARS-CoV-2 calibration curves for each individual assay are straightforward to establish with the appropriate standards. Many are commercially available or have been established by clinical laboratories with an interest in robust understanding of the performance of their SARS-CoV-2 PCR assays.
However, this essential data is rarely reported — some clues appear in limit of detection claims, but very few standard curves are published outside academic literature.
It is frustrating and tragic that the major (perhaps only) advantage of qPCR is ignored. Practices must change to require that a standardized viral load measure is routinely reported to physicians, epidemiologists and patients to inform their critical decisions.




